Using R more "wisely"
Column-wise & row-wise operations with dplyr
Brendan Cullen
dplyr-wisely.netlify.app
About me
![]()
Brendan Cullen
NSF Graduate Research Fellow, University of Oregon
RStudio Certified Instructor
Portland, OR, USA
bcullen.rbind.io | _bcullen | brendanhcullen
About you
✅ Familiar with the tidyverse
✅ Comfortable with common dplyr verbs,
e.g. mutate() and summarize()
✅ Want to more efficiently wrangle
and summarize data with dplyr
1️⃣
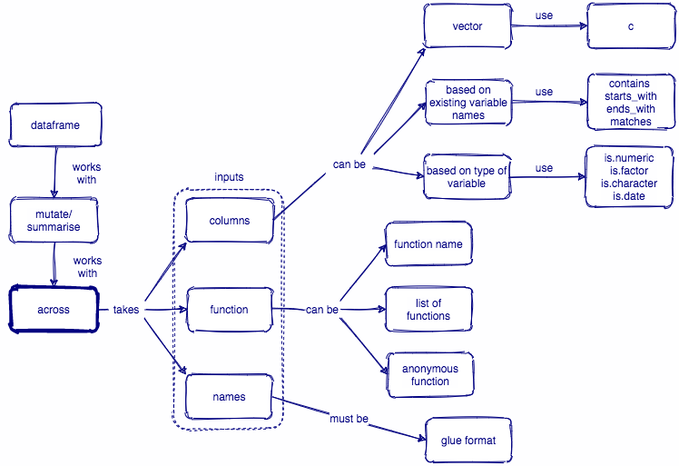
Column-wise operations
Learning objectives
- Review how to apply
dplyrfunctions, e.g.mutate(),summarize(), to single columns
Learn how to perform column-wise operations two ways:
scoped verbs, e.g.
summarize_at(),summarize_if(), etc...across()fromdplyr1.0.0
- Apply
across()to summarize multiple columns of data
Column-wise operations
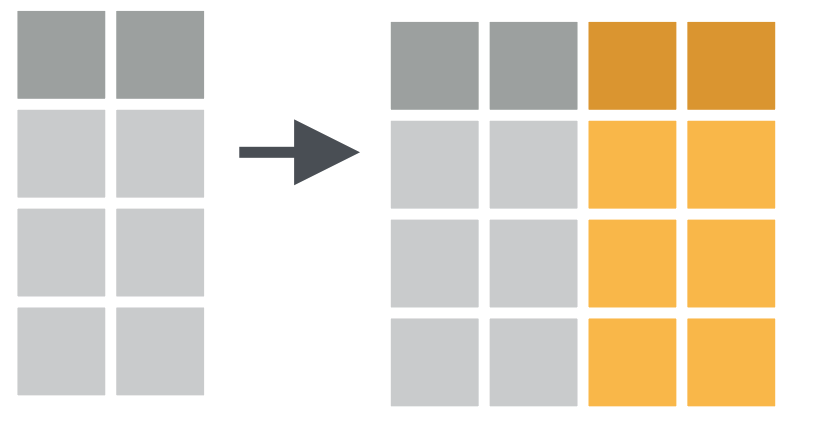
Column-wise operations refers to applying the same dplyr verbs (or other data transformation functions) to multiple columns simultaneously.
Column-wise operations
Column-wise operations refers to applying the same dplyr verbs (or other data transformation functions) to multiple columns simultaneously.
e.g. Create multiple new columns with mutate()
Column-wise operations
Column-wise operations refers to applying the same dplyr verbs (or other data transformation functions) to multiple columns simultaneously.
e.g. Create multiple new columns with mutate()

e.g. Summarize multiple columns with summarize()
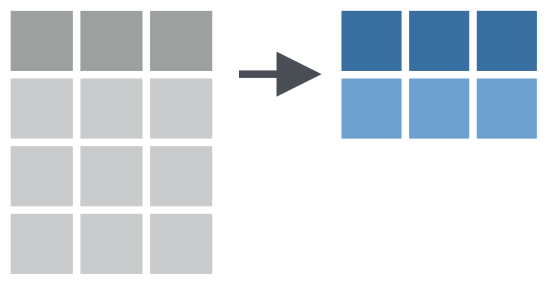
Palmer Penguins
library(palmerpenguins)library(tidyverse)glimpse(penguins)## Rows: 344## Columns: 8## $ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Ade…## $ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgers…## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1,…## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1,…## $ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 18…## $ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475,…## $ sex <fct> male, female, female, NA, female, male, female, mal…## $ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 200…Column-wise operations
Let's review...
Apply summarize() to a single column.

Column-wise operations
Let's review...
Apply summarize() to a single column.

🤔 Use summarize() to calculate the mean bill length for each species in penguins
❓ What function do you need to include before summarize() in order to calculate means for each species?

Artwork by @allison_horst
penguins %>% group_by(species) %>% summarize(bill_length_mm = mean(bill_length_mm, na.rm = TRUE))## # A tibble: 3 x 2## species bill_length_mm## <fct> <dbl>## 1 Adelie 38.8## 2 Chinstrap 48.8## 3 Gentoo 47.5Column-wise operations
Apply summarize() to multiple columns at once

Column-wise operations
Apply summarize() to multiple columns at once

🤔 Calculate the mean bill length and bill depth for each species
🚫 Avoid copying and pasting or repeating mean() more than once in your solution
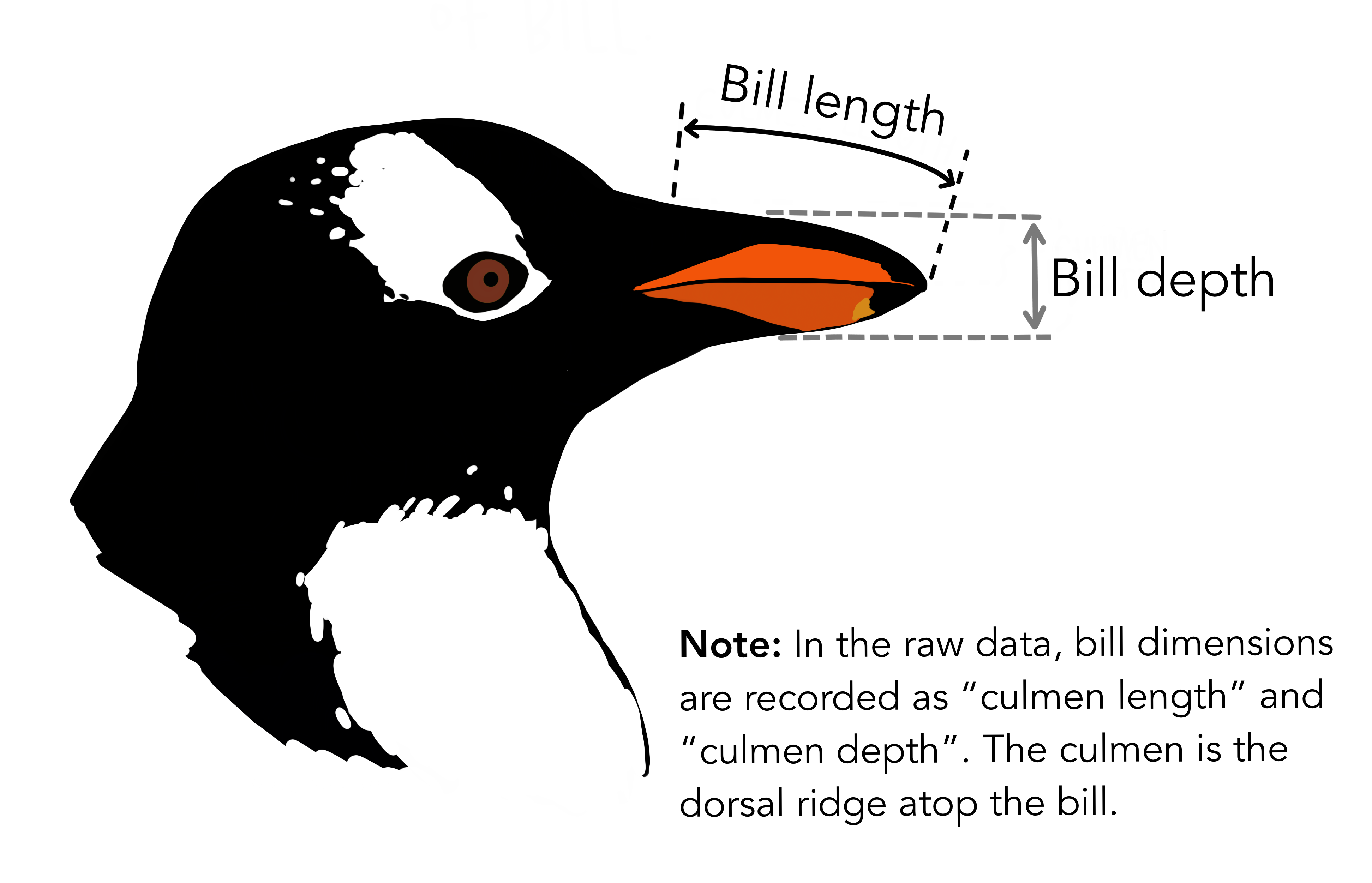
Artwork by @allison_horst
penguins %>%
group_by(species) %>%
summarize_at(vars(c(bill_length_mm, bill_depth_mm)),
mean, na.rm = TRUE)
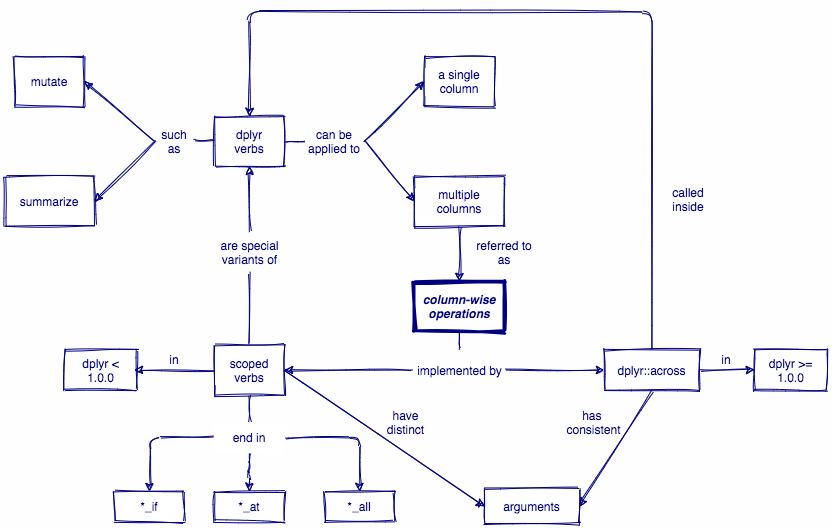
🔍 summarize_at() is an example of a scoped verb. It is a special variant of summarize() that applies a summary function to a specific set of columns.
❗ When using scoped verbs ending in _at() you must use vars() to select columns.
## # A tibble: 3 x 3## species bill_length_mm bill_depth_mm## <fct> <dbl> <dbl>## 1 Adelie 38.8 18.3## 2 Chinstrap 48.8 18.4## 3 Gentoo 47.5 15.0Column-wise operations
Apply summarize() to multiple columns at once

🤔 Calculate the mean of all numeric variables for each species.
🚫 Again, no copying and pasting or repeating mean() more than once!
penguins %>%
group_by(species) %>%
summarize_if(is.numeric, mean, na.rm = TRUE)
🔍 summarize_if() is another example of a scoped verb. It is a special variant of summarize() that applies a summary function to a set of columns that all satisfy some logical criteria.
That logical criteria is specified using a predicate function, e.g. is.numeric(), which will return TRUE or FALSE.
## # A tibble: 3 x 6## species bill_length_mm bill_depth_mm flipper_length_mm body_mass_g year## <fct> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 Adelie 38.8 18.3 190. 3701. 2008.## 2 Chinstrap 48.8 18.4 196. 3733. 2008.## 3 Gentoo 47.5 15.0 217. 5076. 2008.A better way?
dplyr < 1.0.0

summarize_at(), summarize_if(), summarize_all(), mutate_if(), mutate_at(), mutate_all(), ...
😓
A better way?
dplyr < 1.0.0

summarize_at(), summarize_if(), summarize_all(), mutate_if(), mutate_at(), mutate_all(), ...
😓
dplyr >= 1.0.0

across()
😎
dplyr::across()
 Artwork by @allison_horst
Artwork by @allison_horst
dplyr::across()
 Artwork by @allison_horst
Artwork by @allison_horst
dplyr::across()
across(.cols, .fns, ..., .names)
dplyr::across()
across(.cols, .fns, ..., .names)
.cols = the columns you want to transform
dplyr::across()
across(.cols, .fns, ..., .names)
.cols = the columns you want to transform
.fns = the function(s) you want to apply to each of the selected columns
dplyr::across()
across(.cols, .fns, ..., .names)
.cols = the columns you want to transform
.fns = the function(s) you want to apply to each of the selected columns
... = additional arguments for the function(s) specified in .fns (e.g. na.rm = TRUE)
dplyr::across()
across(.cols, .fns, ..., .names)
.cols = the columns you want to transform
.fns = the function(s) you want to apply to each of the selected columns
... = additional arguments for the function(s) specified in .fns (e.g. na.rm = TRUE)
.names = how you want to name the output columns. Here, "{col}" is a special placeholder for the input column name, and you can add any suffix you want to it
dplyr::across()
across(.cols, .fns, ..., .names)
.cols = the columns you want to transform
.fns = the function(s) you want to apply to each of the selected columns
... = additional arguments for the function(s) specified in .fns (e.g. na.rm = TRUE)
.names = how you want to name the output columns. Here, "{col}" is a special placeholder for the input column name, and you can add any suffix you want to it
- e.g. When calculating the mean of
penguins$year, specifying .names = "{col}_mean" would result in an output column named "year_mean"
dplyr::across()
across(.cols, .fns, ..., .names)
🤔 Use across() to calculate the mean of all numeric columns for each species in penguins
📣 Remember, across() goes inside of the dplyr function that you want to apply to multiple columns.
penguins %>%
group_by(species) %>%
summarize(across(where(is.numeric),
mean,
na.rm = TRUE,
.names = '{col}_mean'))
🔍 where() is an example of a tidyselect helper function, like starts_with() or contains().
It selects the variables for which some predicate function, such as is.numeric() returns TRUE.
## # A tibble: 3 x 6## species bill_length_mm_… bill_depth_mm_m… flipper_length_… body_mass_g_mean## <fct> <dbl> <dbl> <dbl> <dbl>## 1 Adelie 38.8 18.3 190. 3701.## 2 Chinst… 48.8 18.4 196. 3733.## 3 Gentoo 47.5 15.0 217. 5076.## # … with 1 more variable: year_mean <dbl>👀 Notice that all of the output variables have the suffix "_mean".
penguins %>%
group_by(species) %>%
summarize(across(where(is.numeric),
list(mean = mean),
na.rm = TRUE))
🔍 If you specify .fns as a named list, e.g. list(mean = mean) , then across() will automatically append the name of the applied function(s) to the names of the output columns.
This way you don't have to manually specify a .names argument at all!
## # A tibble: 3 x 6## species bill_length_mm_… bill_depth_mm_m… flipper_length_… body_mass_g_mean## <fct> <dbl> <dbl> <dbl> <dbl>## 1 Adelie 38.8 18.3 190. 3701.## 2 Chinst… 48.8 18.4 196. 3733.## 3 Gentoo 47.5 15.0 217. 5076.## # … with 1 more variable: year_mean <dbl>👀 Same exact output as before!
dplyr::across()
Multiple summaries with across() -- highly flexible!
penguins %>% group_by(island) %>% summarize( n = n(), across(where(is.factor), n_distinct), across(where(is.numeric), mean, na.rm = TRUE) )## # A tibble: 3 x 9## island n species sex bill_length_mm bill_depth_mm flipper_length_…## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 Biscoe 168 2 3 45.3 15.9 210.## 2 Dream 124 2 3 44.2 18.3 193.## 3 Torge… 52 1 3 39.0 18.4 191.## # … with 2 more variables: body_mass_g <dbl>, year <dbl>More resources
Further reading 📖
Blog post by Hadley Wickham highlighting
across()as a key component of thedplyr1.0.0 update.This vignette on tidyverse.org about column-wise operations.
Two blog posts by Rebecca Barter on scoped verbs and
across().
Practice 💻
Small collection of exercises I put together to accompany these slides
Interactive tutorial by Ted Laderas on
tidyselectand a full list of selection helpers.
05:00
Q & A
2️⃣
Row-wise operations
Learning objectives
- Learn and apply
rowwise()to produce row-by-row summaries
- Use
nest_by()to create list-columns
- Run multiple statistical models on a list-column using
rowwise()
Row-wise operations
rowwise() lets you run operations one row at a time on the data. It’s a lot like group_by(), but it groups the data by row, rather than group.
Row-wise operations
rowwise() lets you run operations one row at a time on the data. It’s a lot like group_by(), but it groups the data by row, rather than group.
What happens when you use rowwise() to create a rowwise data frame? 👀
penguins %>% rowwise()## # A tibble: 344 x 8## # Rowwise: ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g## <fct> <fct> <dbl> <dbl> <int> <int>## 1 Adelie Torge… 39.1 18.7 181 3750## 2 Adelie Torge… 39.5 17.4 186 3800## 3 Adelie Torge… 40.3 18 195 3250## 4 Adelie Torge… NA NA NA NA## 5 Adelie Torge… 36.7 19.3 193 3450## 6 Adelie Torge… 39.3 20.6 190 3650## 7 Adelie Torge… 38.9 17.8 181 3625## 8 Adelie Torge… 39.2 19.6 195 4675## 9 Adelie Torge… 34.1 18.1 193 3475## 10 Adelie Torge… 42 20.2 190 4250## # … with 334 more rows, and 2 more variables: sex <fct>, year <int>Row-wise operations
Let's say we want to compute the mean of bill_length_mm and bill_depth_mm for each penguin.
Row-wise operations
Let's say we want to compute the mean of bill_length_mm and bill_depth_mm for each penguin.
❗ Using mutate() and mean() won't give us what we want.
penguins %>% mutate(avg_bill = mean(c(bill_length_mm, bill_depth_mm), na.rm = TRUE)) %>% select(species, island, avg_bill)## # A tibble: 344 x 3## species island avg_bill## <fct> <fct> <dbl>## 1 Adelie Torgersen 30.5## 2 Adelie Torgersen 30.5## 3 Adelie Torgersen 30.5## 4 Adelie Torgersen 30.5## 5 Adelie Torgersen 30.5## 6 Adelie Torgersen 30.5## 7 Adelie Torgersen 30.5## 8 Adelie Torgersen 30.5## 9 Adelie Torgersen 30.5## 10 Adelie Torgersen 30.5## # … with 334 more rowsRow-wise operations
If we first use rowwise() to create a rowwise data frame, then this calculation will be carried out row-by-row (i.e. for each penguin)
penguins %>% rowwise() %>% mutate(avg_bill = mean(c(bill_length_mm, bill_depth_mm), na.rm = TRUE)) %>% select(species, island, avg_bill)## # A tibble: 344 x 3## # Rowwise: ## species island avg_bill## <fct> <fct> <dbl>## 1 Adelie Torgersen 28.9## 2 Adelie Torgersen 28.4## 3 Adelie Torgersen 29.2## 4 Adelie Torgersen NaN ## 5 Adelie Torgersen 28 ## 6 Adelie Torgersen 30.0## 7 Adelie Torgersen 28.4## 8 Adelie Torgersen 29.4## 9 Adelie Torgersen 26.1## 10 Adelie Torgersen 31.1## # … with 334 more rowsRow-wise operations
c_across() uses tidy selection syntax so you can succinctly select many variables at once
penguins %>% rowwise() %>% mutate(avg_bill = mean(c_across(contains("bill")), na.rm = TRUE)) %>% select(species, island, avg_bill)## # A tibble: 344 x 3## # Rowwise: ## species island avg_bill## <fct> <fct> <dbl>## 1 Adelie Torgersen 28.9## 2 Adelie Torgersen 28.4## 3 Adelie Torgersen 29.2## 4 Adelie Torgersen NaN ## 5 Adelie Torgersen 28 ## 6 Adelie Torgersen 30.0## 7 Adelie Torgersen 28.4## 8 Adelie Torgersen 29.4## 9 Adelie Torgersen 26.1## 10 Adelie Torgersen 31.1## # … with 334 more rowsList columns
Data frames are not limited to atomic vectors. They can also contain recursive vectors, i.e. lists in the form of a list column.
A list column can be a list of data frames.
List columns
Data frames are not limited to atomic vectors. They can also contain recursive vectors, i.e. lists in the form of a list column.
A list column can be a list of data frames.
You can create a list column containing data frames using nest_by().
List columns
Data frames are not limited to atomic vectors. They can also contain recursive vectors, i.e. lists in the form of a list column.
A list column can be a list of data frames.
You can create a list column containing data frames using nest_by().
nested_penguins <- penguins %>% nest_by(species)nested_penguins## # A tibble: 3 x 2## # Rowwise: species## species data## <fct> <list<tbl_df[,7]>>## 1 Adelie [152 × 7]## 2 Chinstrap [68 × 7]## 3 Gentoo [124 × 7]Notice the following 👀
Similar to
group_by()but it visually changes the structure of the dataReturns a rowwise data frame
Default output column name is
data(can override with.keyargument)
List columns
How would you extract the data frame containing the data for Chinstrap penguins out of nested_penguins?
List columns
How would you extract the data frame containing the data for Chinstrap penguins out of nested_penguins?
Notice it does not contain the species variable 👀. By default, .keep = FALSE.
nested_penguins$data[[2]]## # A tibble: 68 x 7## island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year## <fct> <dbl> <dbl> <int> <int> <fct> <int>## 1 Dream 46.5 17.9 192 3500 fema… 2007## 2 Dream 50 19.5 196 3900 male 2007## 3 Dream 51.3 19.2 193 3650 male 2007## 4 Dream 45.4 18.7 188 3525 fema… 2007## 5 Dream 52.7 19.8 197 3725 male 2007## 6 Dream 45.2 17.8 198 3950 fema… 2007## 7 Dream 46.1 18.2 178 3250 fema… 2007## 8 Dream 51.3 18.2 197 3750 male 2007## 9 Dream 46 18.9 195 4150 fema… 2007## 10 Dream 51.3 19.9 198 3700 male 2007## # … with 58 more rowsMany models
You can use a rowwise workflow to run a separate model on each data set in a list column.
Many models
You can use a rowwise workflow to run a separate model on each data set in a list column.
penguins_models <- nested_penguins %>% mutate(model = list(lm(bill_length_mm ~ flipper_length_mm, data = data)))penguins_models## # A tibble: 3 x 3## # Rowwise: species## species data model ## <fct> <list<tbl_df[,7]>> <list>## 1 Adelie [152 × 7] <lm> ## 2 Chinstrap [68 × 7] <lm> ## 3 Gentoo [124 × 7] <lm>Many models
You can use a rowwise workflow to run a separate model on each data set in a list column.
penguins_models <- nested_penguins %>% mutate(model = list(lm(bill_length_mm ~ flipper_length_mm, data = data)))penguins_models## # A tibble: 3 x 3## # Rowwise: species## species data model ## <fct> <list<tbl_df[,7]>> <list>## 1 Adelie [152 × 7] <lm> ## 2 Chinstrap [68 × 7] <lm> ## 3 Gentoo [124 × 7] <lm>❗ Notice that you must wrap your call to lm() inside list() in order for the model output to fit into a list column.
Many models
However, if we access the model column, you’ll notice that it just prints the output of each fit.
penguins_models$model[[1]]## ## Call:## lm(formula = bill_length_mm ~ flipper_length_mm, data = data)## ## Coefficients:## (Intercept) flipper_length_mm ## 13.5871 0.1327Many models
However, if we access the model column, you’ll notice that it just prints the output of each fit.
penguins_models$model[[1]]## ## Call:## lm(formula = bill_length_mm ~ flipper_length_mm, data = data)## ## Coefficients:## (Intercept) flipper_length_mm ## 13.5871 0.1327That output is not super helpful. How can we get more useful information? 🤔
Many models
We can use functions from the broom package to extract summary information about each model in a tidy data frame.
Many models
We can use functions from the broom package to extract summary information about each model in a tidy data frame.
broom::tidy() returns a row for each coefficient in the model. Each column gives information about the estimate or its variability.
nested_penguins %>% mutate(model = list(lm(bill_length_mm ~ flipper_length_mm, data = data))) %>% summarize(broom::tidy(model))## # A tibble: 6 x 6## # Groups: species [3]## species term estimate std.error statistic p.value## <fct> <chr> <dbl> <dbl> <dbl> <dbl>## 1 Adelie (Intercept) 13.6 6.00 2.27 2.49e- 2## 2 Adelie flipper_length_mm 0.133 0.0315 4.21 4.47e- 5## 3 Chinstrap (Intercept) 5.59 9.96 0.562 5.76e- 1## 4 Chinstrap flipper_length_mm 0.221 0.0508 4.34 4.92e- 5## 5 Gentoo (Intercept) -20.7 7.04 -2.94 3.88e- 3## 6 Gentoo flipper_length_mm 0.314 0.0324 9.69 8.60e-17broom::glance() returns a row for each model. Each column gives a model summary: either a measure of model quality, or complexity, or a combination of the two.
nested_penguins %>% mutate(model = list(lm(bill_length_mm ~ flipper_length_mm, data = data))) %>% summarize(broom::glance(model))## # A tibble: 3 x 13## # Groups: species [3]## species r.squared adj.r.squared sigma statistic p.value df logLik AIC## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 Adelie 0.106 0.100 2.53 17.7 4.47e- 5 1 -353. 712.## 2 Chinst… 0.222 0.211 2.97 18.9 4.92e- 5 1 -169. 345.## 3 Gentoo 0.437 0.432 2.32 94.0 8.60e-17 1 -277. 560.## # … with 4 more variables: BIC <dbl>, deviance <dbl>, df.residual <int>,## # nobs <int>More resources
Further reading 📖
Blog post by Hadley Wickham demonstrating helpful use cases for
rowwise(), e.g. simulations and modelingThis vignette on tidyverse.org about row-wise operations.
Chapter 25 from r4ds on "Many models"
Practice 💻
- Interactive tutorial by Ted Laderas on learning
rowwise()
Acknowledgements
Much of this material was based on blog posts written by Hadley Wickham and Rebecca Barter along with tutorials developed by Ted Laderas, which you can access through his tidyowl package.
Thanks also to Allison Horst for the great artwork!